A study published in the journal Nature Commun. by Dr. Carlos Fernández-Tornero's group at the Margarita Salas Center for Biological Research (CSIC) has identified the mechanism that allows certain transposons, also called jumping genes, to insert themselves into safe regions of the genome, which facilitates adaptation to new external conditions without causing dangerous mutations. This study, which is the result of an international collaboration with the CNRS (France), opens the way to improve viral vectors used in gene therapy against diseases such as cancer, hemophilia or congenital blindness.
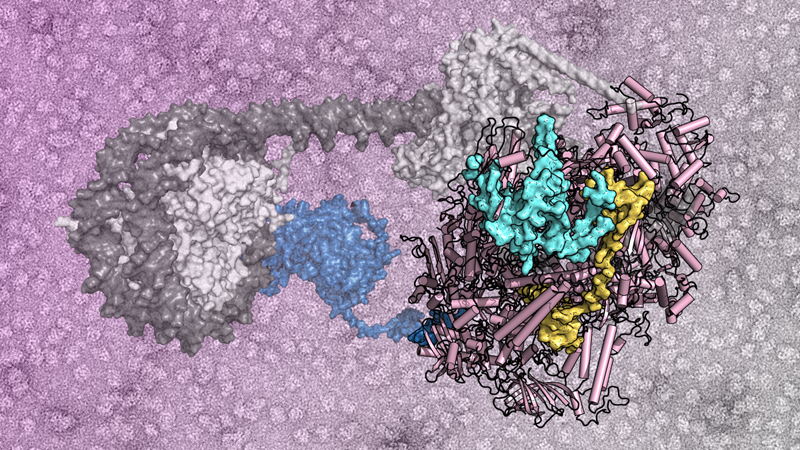
The current work provides details at the atomic level of the interaction between the protein that catalyzes the insertions of the most abundant transposon in beer yeast and RNA polymerase III, an enzymatic complex that binds DNA to synthesize RNA. Such interaction determines the insertion of the transposon into regions of the genome that do not contain essential genes.
"The movement of transposons within the genome allows living organisms to develop new cellular functions and, thus, adapt to different environments, but can pose a threat if it creates harmful mutations," explains Sonia Huecas, one of the lead authors of the study. She adds, "In fact, more than a hundred hereditary diseases have been attributed to new transposon insertions."
"Using electron cryomicroscopy, we describe how an unfolded region of the transposon insertion protein docks into a crevice on the surface of RNA polymerase III," says Carlos Fernández Tornero, who co-directed the study with Pascale Lesage of the Institut de Recherche Saint Louis. "But what surprised us most is that this interaction reconfigures RNA polymerase III so that it is trapped longer on the DNA and thus increases the chances of insertion of the transposon into safe places in the yeast genome," adds Tornero.
In addition to being a breakthrough for basic research, this study may help to improve viral vectors used in gene therapy, which are currently integrated into gene-rich regions, where they can have harmful effects. The researchers plan to use the new atomic information about the interaction to design a motif capable of directing gene therapy vectors to safe regions of the genome and, thus, limiting their mutagenic potential.
Reference: Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration. Phong Quoc Nguyen, Sonia Huecas, Amna Asif-Laidin, Adrián Plaza-Pegueroles, Beatrice Capuzzi, Noé Palmic, Christine Conesa, Joël Acker, Juan Reguera, Pascale Lesage & Carlos Fernández-Tornero (2023). Nat Commun 14, 1729. https://doi.org/10.1038/s41467-023-37109-4
More information:
CSIC Press Release (in Spanish): link.

