The research group led by Dr. M. Cristina Vega at the CIB Margarita Salas has determined the structure of a new virulence factor of Leptospira interrogans, the bacterium responsible for leptospirosis, the most widespread zoonotic disease. This work, published in Frontiers in Immunology in collaboration with the group of Dr. Santiago Rodríguez de Córdoba, delves into the immunoevasive mechanisms of Leptospira through the inhibition of the signaling pathways and neutrophil recruitment of anaphylatoxin C5a during the Infective process.
Leptospirosis is a neglected zoonotic disease caused by the Gram-negative spirochete Leptospira. This bacterium colonizes humans, domestic and farm animals, and wild species such as mice, rats, and bats, which serve as reservoirs of infection. In almost all of the one million cases of leptospirosis that are detected each year in humans, the disease causes mild to severe symptoms that can result in fatality. Climate change is also accelerating the spread of the pathogen and increasing the number of annual cases of leptospirosis.
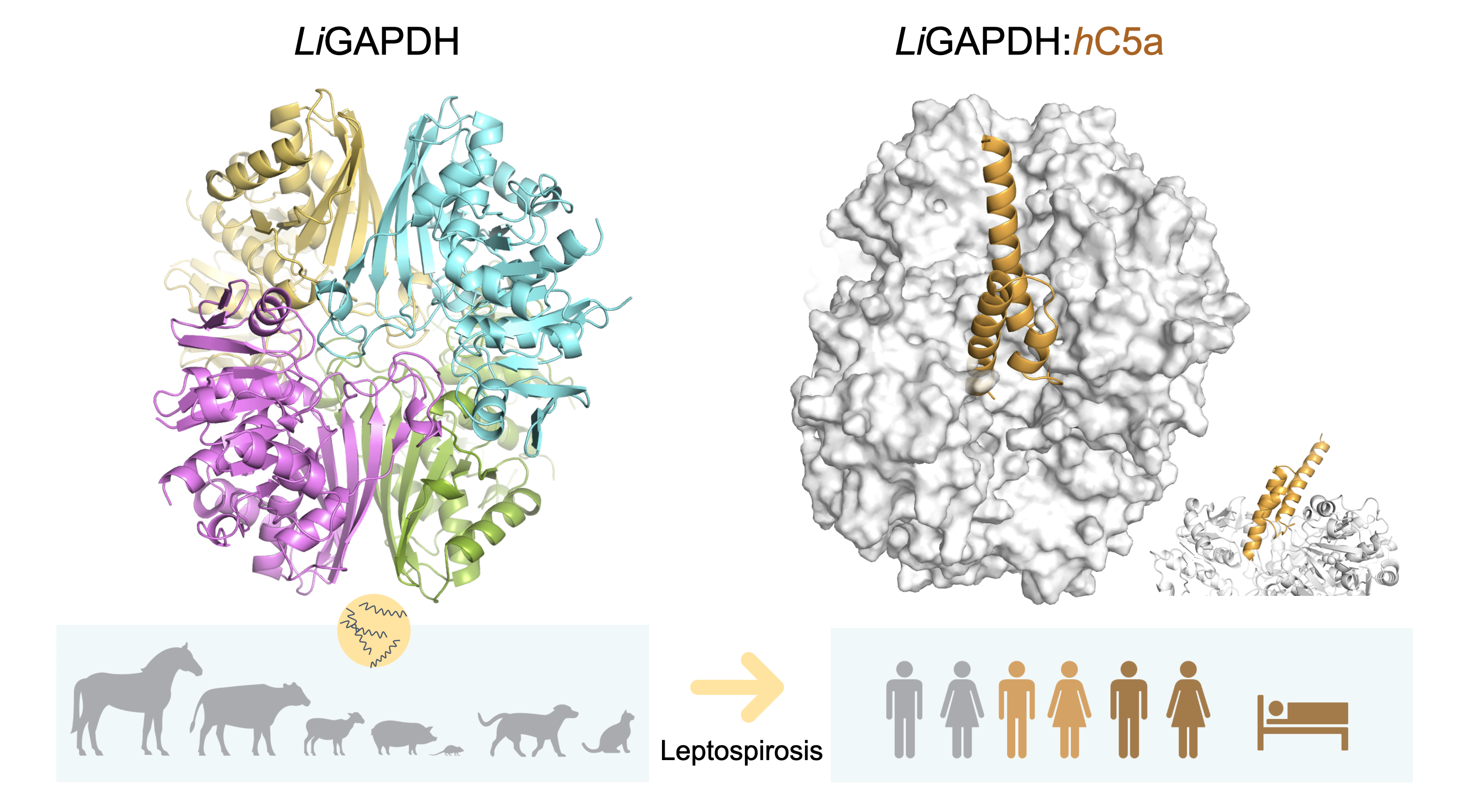
Leptospira presents multiple factors involved in defense mechanisms, to which this work adds a glycolytic enzyme with moonlighting functions, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH is one of the most abundant proteins in the extracellular proteome, where it can act as a virulence factor and enhance infectivity and evasion of the immune system.
The structure of LiGAPDH by X-ray crystallography at 2.37 Å provides a detailed description of this key protein in physiological and pathological processes, and helps to understand its immune evasive role as a virulence factor and its possible applications in biomedicine. In this study, in addition to characterizing the kinetic parameters of the enzyme on the natural substrates, it has been established by cross-linking and biolayer interferometry that LiGAPDH can interact in vitro with the anaphylatoxin C5a of the complement system, with a binding mode characterized by slow association and dissociation constants. Docking studies guided by cross-linking experimental results show that the bundle of α-helices characteristic of the anaphylatoxin structure, with the only free cysteine residue, interacts with the area of the active site of LiGAPDH responsible for binding to the substrate glyceraldehyde 3-phosphate (G3P) in the N-terminal domain. The buried surface area of the interaction is ~3,000 Å2 and the nature of the interaction is predominantly polar, where the bundle of α-helices of C5a has a negative charge and the active center of LiGAPDH has a positive charge. This mode of interaction is consistent with other GAPDH complexes with α-helical proteins. Since GAPDH is a highly conserved protein, the binding model can be extrapolated to many other pathogenic microorganisms in which GAPDH acts as a virulence factor by sequestering C5a.
The present work adds L. interrogans to the growing list of bacterial pathogens in which glycolytic enzymes such as GAPDH act as immunoevasion factors. The groups of Dr. Vega and Dr. Rodríguez de Córdoba belong to the CSIC Global Health Platform where they collaborate in studies of infectious and inflammatory processes in which C5a is involved. In recent years, in collaboration with the groups led by José Ramón Regueiro from the UCM and Sebastián Albertí Serrano from the UIB, it has been established that the sequestration of the anaphylatoxin C5a by GAPDH significantly inhibits the chemoattraction of neutrophils by C5a. In this way, pathogens such as Atopobium vaginae or Streptococcus pyogenes, and now L. interrogans, reduce the response of the innate immune system and favor the infectious process.
These results are part of the Thematic Research Network of Complement in Health and Disease (RED2022-134750-T) and have been funded by the Biomedicine Consortium of the Comunidad de Madrid Complemento III-CM (P2022/BMD-7278).
Reference: The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity. Navas-Yuste S, de la Paz K, Querol-García J, Gómez S, Rodríguez de Córdoba S, Fernández FJ, Vega MC [2023]. Front Immunology 14:1190943. https://doi.org/10.3389/fimmu.2023.1190943
More information:
- Gómez S, Querol-García J, Sánchez-Barrón G, Subías M, González-Alsina A, Franco-Hidalgo V, Albertí S, Rodríguez de Córdoba S, Fernández FJ, Vega MC [2019]. Front Microbiol. 10:326. https://www.frontiersin.org/articles/10.3389/fmicb.2019.00326/full
- Fernández FJ, Gómez S, Vega MC [2019]. Seminars Cell Dev Biol. 85, 98-109. https://www.sciencedirect.com/science/article/abs/pii/S1084952117301386?via%3Dihub
- Querol-García J, Fernández FJ, Marin AV, Gómez S, Fullà D, Melchor-Tafur C, Franco-Hidalgo V, Albertí S, Juanhuix J, Rodríguez de Córdoba S, Regueiro JR, Vega MC. [2017]. Front Microbiol 8:541. https://www.frontiersin.org/articles/10.3389/fmicb.2017.00541/full

