One of the mechanisms that determine how cells enter latency when nutrients lack has been revealed in a paper published at eLife by scientists at the Center for Biological Research (CIB-CSIC), in collaboration with the Institute of Functional Biology (CSIC-USAL) in Salamanca and the IRB Barcelona.
RNA polymerase I is the enzyme that synthesizes the molecular machinery responsible for producing all the proteins in the cell. This enzyme functions at a very high speed when cells grow but its inactivation is essential when cells stop growing. However, in many cancer cells, the activity of this enzyme uncontrollably increases to expand the tumor.
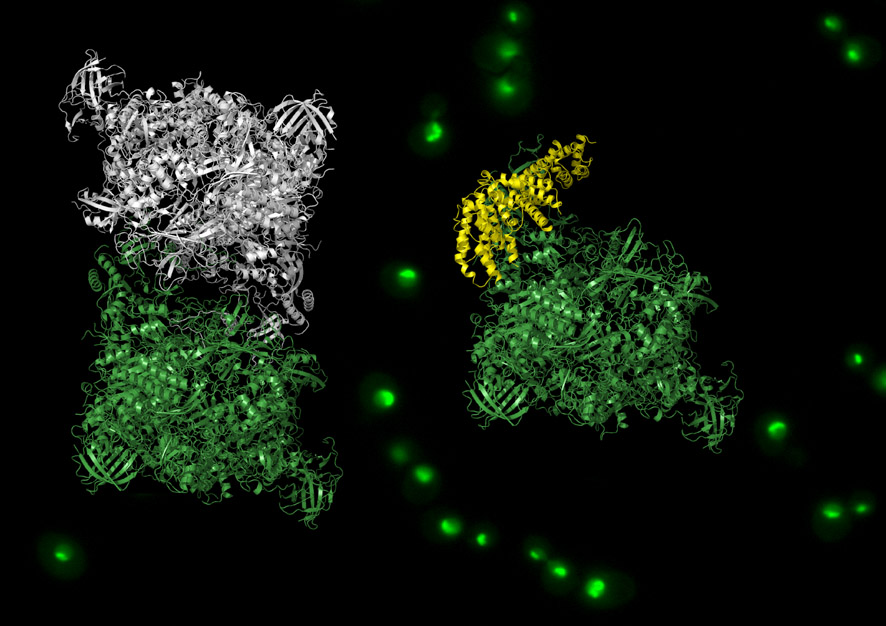
The CIB group led by Carlos Fernández Tornero determined in 2013 the atomic structure of RNA polymerase I in its inactive state. They have now been able to understand how this enzyme is activated and inactivated by combining new structural studies with advanced techniques of molecular analysis, genetic engineering and live cell microscopy, in collaboration with research teams at the IBGF and the IRB.
These researchers show that cells form different assemblies of RNA polymerase I according to nutrients availability: upon starvation two copies of this enzyme bind each other and they mutually inactivate, whereas when nutrients are unlimited RNA polymerase I is freed to bind activating proteins that promote the synthesis of new proteins.
The discovery of this mechanism opens new ways to control cell growth, which has obvious implications for cancer research.
Reference: E. Torreira, J.A. Louro, I. Pazos, N. González-Polo, D. Gil-Carton, A.G. Duran, S. Tosi, O. Gallego, O. Calvo, C. Fernández-Tornero. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. eLife DOI: 10.7554/eLife.20832
Other related links:

