An article published in the International Journal of Biological Macromolecules by the Biomolecular NMR group of the Margarita Salas Center for Biological Research (CSIC), led by Dr. Francisco J. Blanco, describes the structure of the ING3 protein and its interaction with histone H3. The work contributes to clarify the molecular mechanisms that might explain the possible deleterious effects of ING3 mutations detected in tumors.
The genome, the information carrier for life, is found in the cell nucleus bound to various proteins forming the chromatin. Most of these proteins are histones, which organize the DNA in a more or less compact form. The degree of compactness (and thus its accessibility) depends on modifications in the histones. Proteins of the ING (INhibitors of Growth) family bind to chromatin and specifically recognize histone H3 methylation at the K4 position. In this way, they recruit enzymes that produce additional histone modifications (acetylation or deacetylation). The set of histone modifications (as well as modifications on the DNA itself) determines to what extent genes are expressed, and an abnormal pattern of modifications can cause pathologies such as cancer.
ING3 is the least studied of the five human ING proteins. Although it is necessary for embryonic development, its excess stimulates cell proliferation and correlates with poor prognosis in prostate cancer patients.
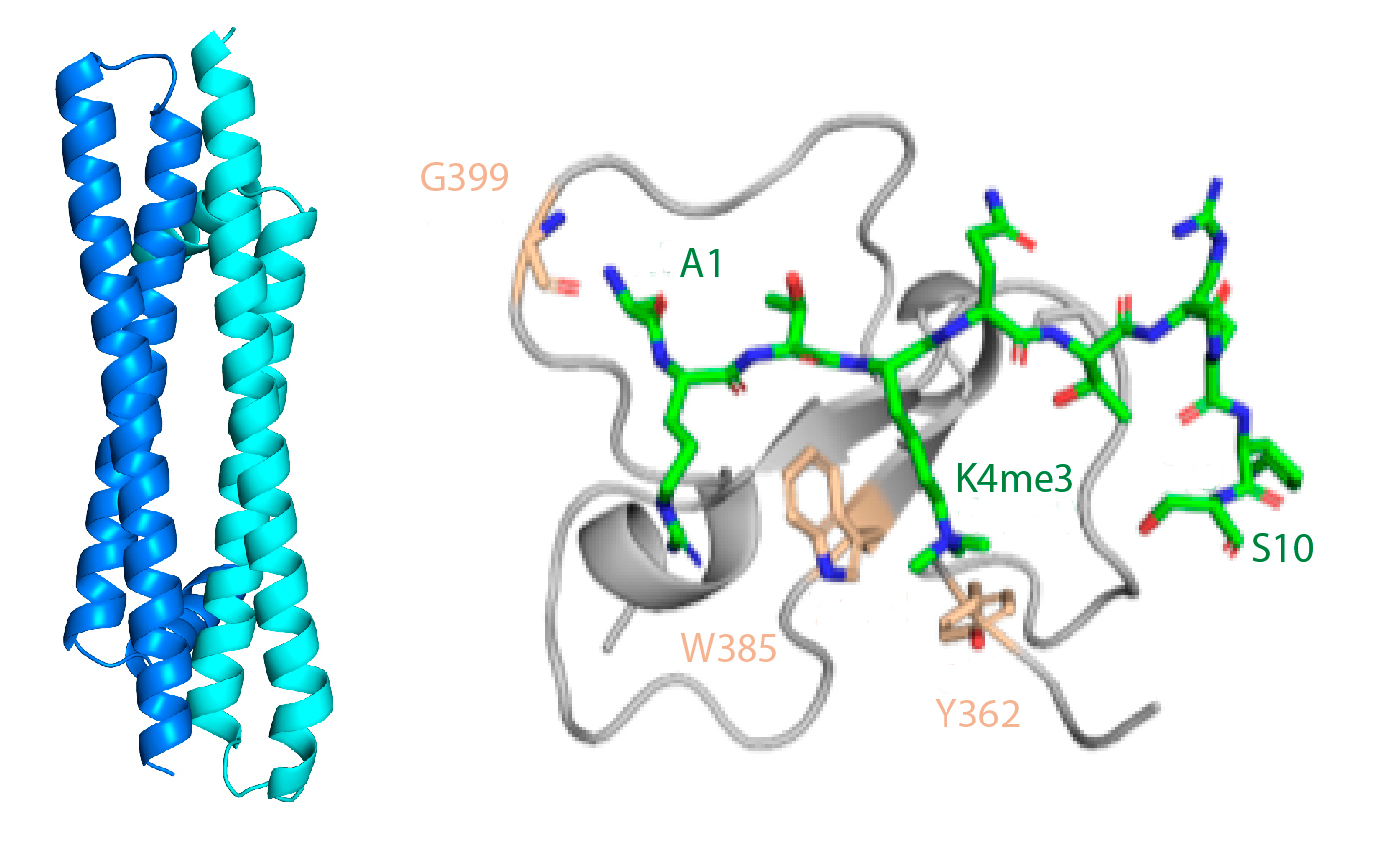
Using NMR and crystallography, Ferreras-Gutiérrez et al. have found that two ING3 molecules assemble into a coiled-coil helical structure with an antiparallel orientation. A long disordered central region separates the helical region from the histone-binding region, making ING3 a bivalent reader of histone H3 methylation, thus increasing the affinity of the interaction. This structure explains why certain mutations in ING3 appear highly frequently in tumors. Some of them destabilize its structure and others interfere with histone recognition.
The structural organization of ING3 is very similar to that of its homologs ING4 and ING5. In the case of ING1 and ING2, their structure is not yet known in detail. Although ING1 and ING2 are tumor suppressors, ING3 is an oncoprotein, whereas ING4 and ING5 cannot be clearly classified into one of those two categories. Further structural and functional studies will be necessary to clarify these and other unknown aspects of the ING protein family.
Reference: Structural analysis of ING3 protein and histone H3 binding. Mariola Ferreras-Gutiérrez, Belén Chaves-Arquero, Amaia González-Magaña, Nekane Merino, Ignacio Amusategui-Mateu, Sonia Huecas, Francisco J. Medrano, Francisco J. Blanco (2023) International Journal of Biological Macromolecules, Vol 242, Part 1, 124724. https://doi.org/10.1016/j.ijbiomac.2023.124724.

