The discovery that the coding sequences of most eukaryotic genes are interrupted by introns led to the study of the molecular mechanisms controlling their cleavage and the formation of mature messengers. Several works have revealed that the spliceosome, a ribonucleoproteic complex constituted by five snRNP and more than 300 auxiliary proteins, through a process known as splicing, drives intron cleavage. In the last decades, it has been demonstrated that this complex plays an essential role in many physiological processes in eukaryotes by ensuring proper gene expression profiles in response to both internal and external stimuli.
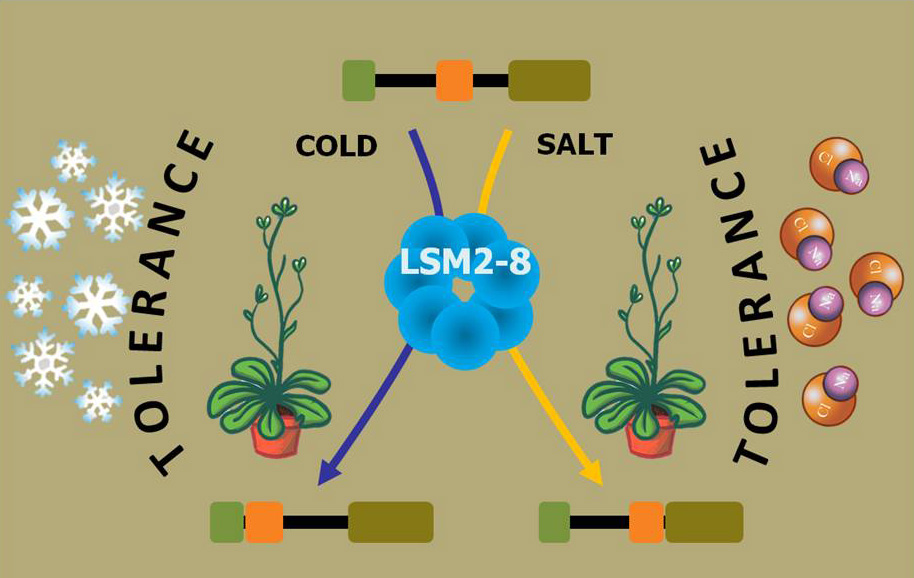
Using the model plant Arabidopsis thaliana, a recent article published by the group of Prof. Julio Salinas in Nucleic Acids Research provides a new perspective on the mechanisms underlying the spliceosome activity. So far, it was assumed that spliceosome activity and specificity was mainly modulated by the action of its auxiliary proteins. This article reports results that break with this paradigm, demonstrating that the LSM2-8 complex from the U6 snRNP, a core component of the spliceosome, determines the specificity of its activity. It is shown that this complex controls the constitutive and alternative splicing of selected transcripts depending on the environmental conditions. More interestingly, the selective role of the complex has relevant physiological implications since it is required for adequate plant adaptation to abiotic stresses.
All these findings unveil an unanticipated function for the LSM2–8 complex that represents a new layer of posttranscriptional regulation in response to external stimuli in eukaryotes.
Reference: Environment-dependent regulation of spliceosome activity by the LSM2-8 complex in Arabidopsis. Cristian Carrasco-Lopez, Tamara Hernandez-Verdeja, Carlos Perea-Resa, David Abia, Rafael Catala and Julio Salinas. Nucleic Acids Research, 2017. doi: 10.1093/nar/gkx375

