Group Leader/s
Group PI/s
intro
Our research program integrates biochemistry, biophysics, and bottom-up synthetic biology to reconstruct minimal bacterial division machineries from its molecular building blocks in controlled cell-like environments. This effort, which is framed on the quest of building synthetic cells from scratch, will contribute to our understanding of how cells divide and will provide new horizons for biotechnological and biomedical applications.
For more details, please see the personal lab web page at https://mastermcib.wixsite.com/grlabcib
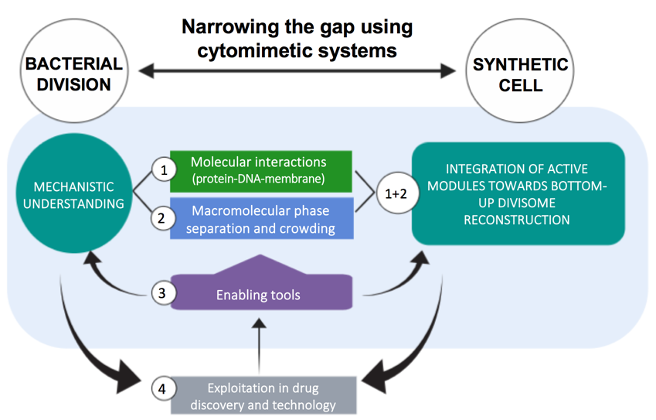
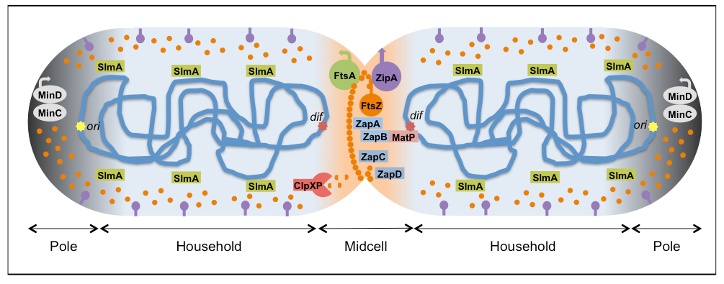
- We aim at understanding the molecular mechanisms responsible for the biochemical organization of the bacterial division machinery (the divisome). The purpose is to achieve a quantitative description of how FtsZ (the central element of the divisome in most bacteria) and the proteins modulating division ring stability work together as an integrated system of multiple interactions to assemble the functional molecular engine that drives cytokinesis. This biochemical information will contribute to defining more precise conditions to reconstruct different combinations of divisome subsets and to test their functional organization in minimal membrane systems and cell-like containers, as vesicles and droplets produced by microfluidics.
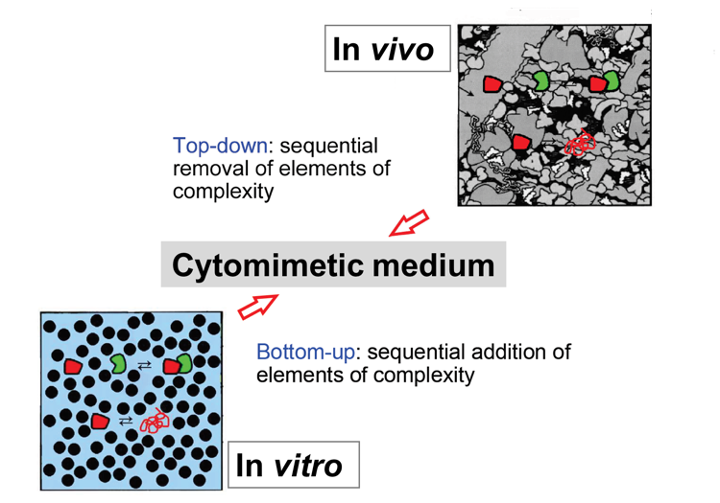
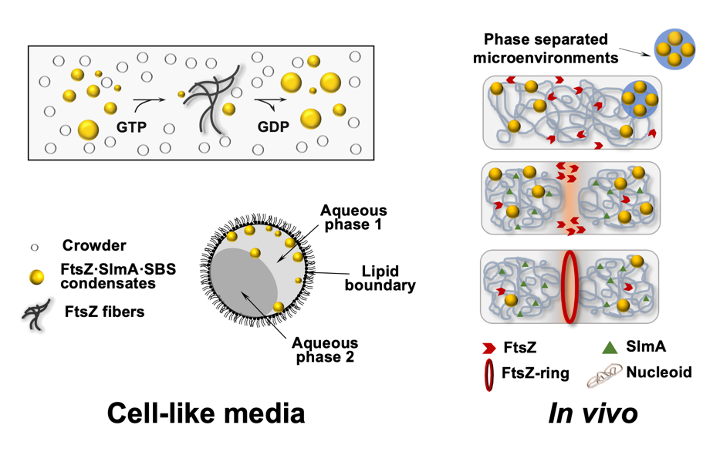
- Synthetic systems can be devised to mimic elements of the intracellular complexity (as excluded volume effects due to natural crowding, surface interactions, and macromolecular condensation resulting from biologically regulated liquid-liquid phase separation) in media of known and controllable composition.
- This integrative approach has allowed us to demonstrate that FtsZ, together with its nucleoid-associated inhibitor SlmA, assembles into dynamic condensates driven by phase separation in crowded cell-like conditions. Our findings suggest that phase-separated biomolecular condensation may also help to organize intracellular space in bacteria.
Selected Publications
Monterroso B, Margolin W, Boersma AJ, Rivas G, Poolman B, Zorrilla S. [2024]. Macromolecular Crowding, Phase Separation, and Homeostasis in the Orchestration of Bacterial Cellular Functions. Chem Rev. 124(4):1899-1949. doi: 10.1021/acs.chemrev.3c00622.
Monterroso B, Robles-Ramos MÁ, Sobrinos-Sanguino M, Luque-Ortega JR, Alfonso C, Margolin W, Rivas G, Zorrilla S. [2023]. Bacterial division ring stabilizing ZapA versus destabilizing SlmA modulate FtsZ switching between biomolecular condensates and polymers. Open Biol.13(3):220324. doi: 10.1098/rsob.220324.
Rivas G, Minton AP. [2022]. Influence of Nonspecific Interactions on Protein Associations: Implications for Biochemistry In Vivo. Annu Rev Biochem. 91:321-351. doi: 10.1146/annurev-biochem-040320-104151.
Robles-Ramos MÁ, Margolin W, Sobrinos-Sanguino M, Alfonso C, Rivas G, Monterroso B, Zorrilla S. [2020]. The Nucleoid Occlusion Protein SlmA Binds to Lipid Membranes. mBio. 11(5):e02094-20. doi: 10.1128/mBio.02094-20.
Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Robles-Ramos MA, López-Álvarez M, Margolin W, Keating CD, Rivas G. [2019]. Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 20(1). pii: e45946. doi: 10.15252/embr.201845946.
Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Robles-Ramos MÁ, Alfonso C, Söderström B, Meiresonne NY, Verheul J, den Blaauwen T, Rivas G. [2019]. The Bacterial DNA Binding Protein MatP Involved in Linking the Nucleoid Terminal Domain to the Divisome at Midcell Interacts with Lipid Membranes. MBio. 10(3). pii: e00376-19. doi: 10.1128/mBio.00376-19.
Ramirez-Diaz DA, García-Soriano DA, Raso A, Mücksch J, Feingold M, Rivas G, Schwille P. [2018]. Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol. 16(5):e2004845. doi: 10.1371/journal.pbio.2004845.
Sobrinos-Sanguino M, Zorrilla S, Keating CD, Monterroso B, Rivas G. [2017]. Encapsulation of a compartmentalized cytoplasm mimic within a lipid membrane by microfluidics. Chem Commun (Camb). 53(35):4775-4778. doi: 10.1039/c7cc01289f.
Rivas G, Minton AP. [2016]. Macromolecular Crowding In Vitro, In Vivo, and In Between. Trends Biochem Sci. 41:970-981.
Martos A, Jiménez M, Rivas G*, Schwille P* [2012]. Towards a bottom-up reconstitution of bacterial cell division. Trends Cell Biol. 22:634-643
Funding
- 2024-2027: MINCELL - The physical basis of cell division in minimal and synthetic cells. The "Fundamentos" Research Program of BBVA Foundation.
- 2023-2027: DIVINOCELL - Reconstituting division in minimal cell-like systems. Spanish Government Plan Nacional I+D+I, PID2022-136951NB-I00.
- 2020-2023: BASYC - Bacterial division in synthetic cytomimetic environments. Spanish Goverment. Plan Nacional I+D+i, PID2019-104544GB-I00.
- 2017-2019: BIOROOMS - Analysis, synthesis and reassembly of the biorooms active in bacterial division. Spanish Government Plan Nacional I+D+i, BFU2016-75471-C2-1-P.
- 2015-2019: MASSTRPLAN - "MASS Spectrometry TRaining network for Protein Lipid adduct Analysis" EU Project 675132 (H2020-MSCA-ITN-2015). Innovative Training Network.
- 2015-2016: ODIVITUBE - Biochemical organization of minimal divisomes in the test tube. Spanish Government, Plan Nacional I+D+i, BFU2014-28941-C03-02.
- 2012-2014: SYNVISION - Synthetic biology of bacterial cell division: reconstruction of minimal divisomes in biomimetic membrane systems. Spanish Government, Plan Nacional I+D+i, BIO2011-28941-C03-03.
- 2011-2014: SYNTHETIC BIOLOGY OF BACTERIAL CELL DIVISION. Human Frontier Science Program RGP0050-2010.
- 2009-2013: DIVINOCELL - Exploiting Gram-negative cell division targets in the test tube to obtain anti-microbial compounds. European Commission, FP7-HEALTH-F3-2009-223431.
More info
GR LAB
Centro de Investigaciones Biológicas Margarita Salas - CSIC
Ramiro de Maeztu 9 - E28040 Madrid
Tel. +34 918373112 ext. 4304
E-mail: grivas@cib.csic.es
GRLAB web: http://www.cib.csic.es/en/grupo.grivas & https://mastermcib.wixsite.com/grlabcib
HOW TO REACH GRLAB:
http://www.cib.csic.es/es/como_llegar.php?
GR lab web: Bioquímica de sistemas de la división bacteriana


