Adaptive gene expression changes of plants after exposure to spaceflight related environments
Space research keeps being a top priority for the more important economies in the world, with emerging agencies as China, India or our Spanish Space Agency (AEE), even after the COVID and war in Ukraine and Gaza context. The next step in space colonization, to visit again the Moon and soon the arrival of the first human to Mars from the Gateway platform, is ready to begin. In this context, the need of space agriculture to sustain life out of Earth is demanding. To understand the molecular mechanisms that allow plants to adapt to the cultivation conditions in orbit or at Moon g-levels is crucial, even with key alterations at the root meristem level of the plants.

The SOS Microgravity Project take advance of CSIC 20 years’ experience in spaceflight, partial gravity and simulated microgravity experiments, to unravel the two fundamental problems of Space Omics research as described by the GeneLab Plant AWG, ISSOP and Space Omics TT international consortia we are coordinating at the European level. The first one is to obtain reproducible, abundant and good quality samples without the constrains imposed by spaceflight experimentation, and the second is to provide new tools that help integrating two decades of spaceflight results to produce a real advance in our global knowledge of life support systems out of Earth.
On the one hand, the SOS Microgravity Project aims to gather new plant samples exposed to complex environmental conditions of interest for Space Exploration. We will perform the Space Omics analysis as part of ESA multidisciplinary experiments including SEEDLING GROWTH (ESA/NASA spaceflight experiment series), ground based simulation facilities projects ROOTROPS and GIA2 (to simulate Mars Environments increased radiation and reduced gravity) and the recently approved PRIMO project (Idea I-2022-04105) executed in the Moon surface.
On the other hand, we will use new bioinformatic approaches to generate, transfer and communicate space omics knowledge to the Society. Firstly, , to develop the DEEP (Degenerate Energy Expression Profiles) transcriptional analysis to reanalyze the –omics datasets from our group (which metadata information is accessible to us). Secondly, a gamification strategy applied in the creation of the “SOS: Microgravity!” mobile app. It will allow us to share Space Omics knowledge with the society, taken advance of public interest for Space Exploration and Mobile Apps combined. More than that, younger gamers will be more engaged to STEM careers by this, and could choose if they help us creating new in-game knowledge, that we will supervise and cross-reference, or they prefer to “buy” in-game research, so they will be funding us to boost the humanity desires of Space Exploration. We are constantly looking for programmers interested into joint the SOS Microgravity Project.
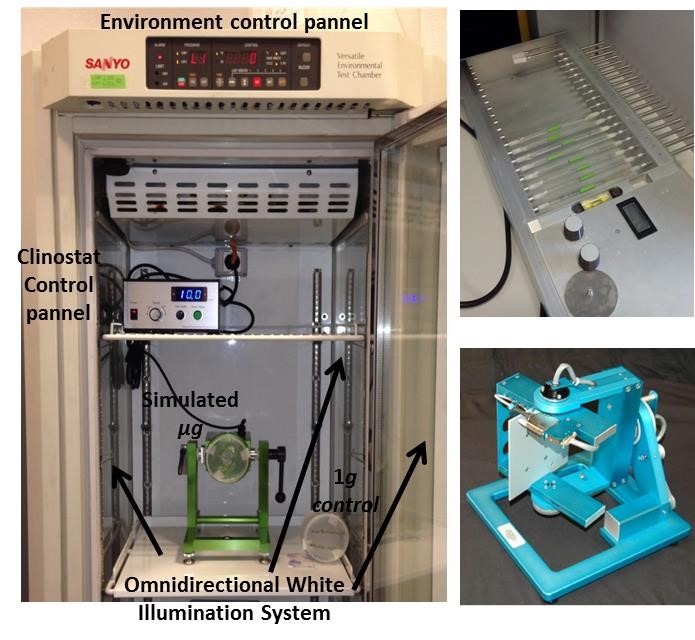
Finally, we actively collaborate with companies developing ground-based simulators and provide access for researchers interested in our simulation facilities in Madrid (clinostat top and U-grav down), and keep leading well-established research teams, international working teams and international committees (NASA/ESA/ELGRA).

Barker, R., Kruse, C.P.S., Johnson, C. et al. (2023) Meta-analysis of the space flight and microgravity response of the Arabidopsis plant transcriptome. npj Microgravity 9, 21 https://doi.org/10.1038/s41526-023-00247-6
Deane CS, Space Omics Topical T, da Silveira WA, Herranz R (2022) Space omics research in Europe: Contributions, geographical distribution and ESA member state funding schemes. iScience 25 (3):103920. doi:10.1016/j.isci.2022.103920
Herranz R, Benguria A, Lavan DA, Lopez-Vidriero I, Gasset G, Javier Medina F, van Loon JJWA, Marco R (2010) Spaceflight-related suboptimal conditions can accentuate the altered gravity response of Drosophila transcriptome. Molecular Ecology 19 (19):4255-4264. doi:10.1111/j.1365-294X.2010.04795.x
Herranz R, Vandenbrink JP, Villacampa A, Manzano A, Poehlman WL, Feltus FA, Kiss JZ, Medina FJ (2019) RNAseq Analysis of the Response of Arabidopsis thaliana to Fractional Gravity Under Blue-Light Stimulation During Spaceflight. Frontiers in plant science 10:1529. doi:10.3389/fpls.2019.01529
Madrigal P, Gabel A, Villacampa A, Manzano A, Deane CS, Bezdan D, Carnero-Diaz E, Medina FJ, Hardiman G, Grosse I, Szewczyk N, Weging S, Giacomello S, Harridge S, Morris-Paterson T, Cahill T, Silveira WAd, Herranz R (2020) Revamping Space-omics in Europe. Cell Systems: https://doi.org/10.1016/j.cels.2020.1010.1006
Manzano A, Villacampa A, Sáez-Vasquez J, Kiss JZ, Medina FJ, Herranz R (2020) The importance of Earth reference controls in spaceflight -omics research: characterization of nucleolin mutants from the Seedling Growth experiments iScience 23 (11):101686. doi:10.1016/j.isci.2020.101686
Rutter L, Barker R, Bezdan D, Cope H, Costes SV, Degoricija L, Fisch KM, Gabitto MI, Gebre S, Giacomello S, Gilroy S, Green SJ, Mason CE, Reinsch SS, Szewczyk NJ, Taylor DM, Galazka JM, Herranz R, Muratani M (2020) A New Era for Space Life Science: International Standards for Space Omics Processing (ISSOP). Patterns:https://doi.org/10.1016/j.patter.2020.100148
(*) Open Access publications available at
Cell Press special issue 2020 https://www.cell.com/c/the-biology-of-spaceflight,
Cell Press special issue 2022 https://www.sciencedirect.com/journal/iscience/special-issue/10K54CFNC0X
Nature journals in 2023 https://www.nature.com/collections/cfhdahhcde.
Space Omics Topical Team, funded by ESA (contract #4000131202/20/NL/PG/pt)
GIA2 Ground Based Facilities project, funded by ESA (contract #4000130341/20/NL/PG/pt)
Research Grant PID2023-150842OB-I00 funded by MCIN/AEI /10.13039/501100011033 /10.13039/501100011033 and by ERDF/EU.

