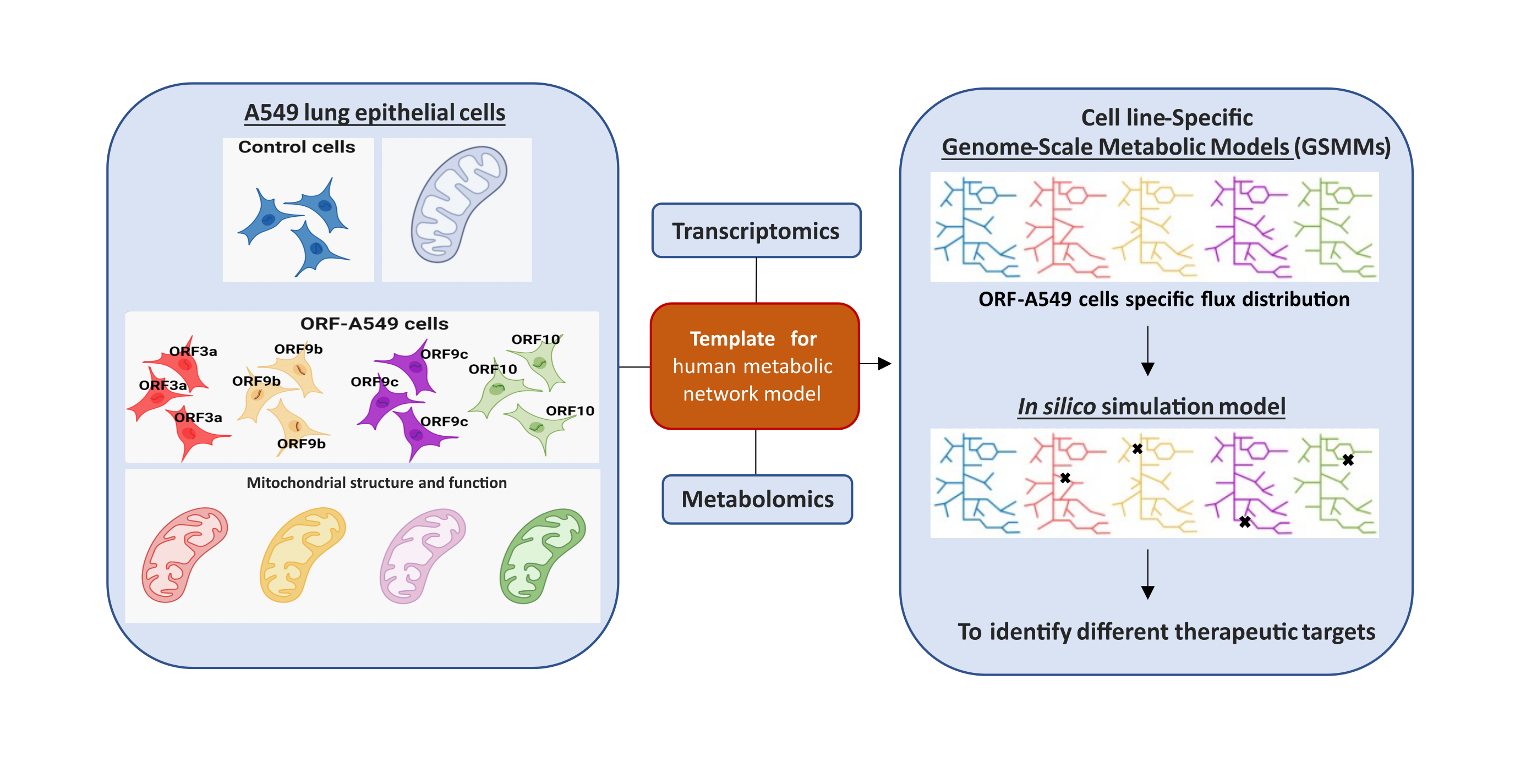
A study published in the Journal of Medical Virology by the group led by María Montoya at the Margarita Salas Biological Research Center (CSIC) demonstrates the impact of accessory proteins of the SARS-CoV-2 virus on the metabolism and mitochondrial function of lung epithelial cells. The study describes the role of four of these proteins, ORF3a, ORF9b, ORF9c, and ORF10, in the metabolic reprogramming of the lung cells. Additionally, it shows the potential of the GSMM bioinformatics model to investigate the pathogenesis of the virus causing COVID-19.
Among the lesser-known proteins of the SARS-CoV-2 virus are 11 so-called accessory proteins that, although not directly related to viral replication and the formation of new viral particles, do appear to be involved in immune evasion strategies by the virus, interactions with metabolic pathways, and induction of cell death. Among them, ORF3a, ORF9b, ORF9c, and ORF10 were selected to study their effect on epithelial cells of pulmonary origin (A549).
López-Ayllón et al. generated cell lines transduced with each of these proteins to study independently the cellular response induced by them using transcriptomic and metabolomic analyses, complemented with various functional assays. In addition, a bioinformatics model known as the Genome-Scale Metabolic Model (GSMM) was used. The application of this model to study the impact of accessory proteins on the cellular metabolism of the host cell is an innovative contribution of this study, as it had previously only been carried out with the whole virus.
The results revealed altered metabolic fluxes in lipid, carbohydrate, protein, and nucleotide metabolism pathways. ORF3a expression mostly altered lipid metabolism pathways, whereas ORF9b, ORF9c, and ORF10 had a greater effect on amino acid metabolism.
In silico simulation models to analyze the modulation of certain molecules or metabolites that could reverse the observed metabolic alteration allowed the researchers to identify different therapeutic targets. Among the targets found for ORF3a-transduced cells, the model was validated with a phospholipase D2 (PLD2) inhibitor. After treatment of the cells with this inhibitor, the mitochondrial and metabolic phenotype of the cells was recovered, resembling that observed in control cells.
The methodology presented in this work highlights the usefulness of GSMMs for analyzing cells transduced only with a viral protein, which could become a valid tool for investigating both the pathogenesis of the SARS-CoV-2 virus and the search for new antiviral strategies.
Reference: Metabolic and mitochondria alterations induced by SARS-CoV-2 accessory proteins ORF3a, ORF9b, ORF9c and ORF10. Blanca D. López-Ayllón, Silvia Marin, Marco Fariñas Fernández, Tránsito García-García, Raúl Fernández-Rodríguez, Ana de Lucas-Rius, Natalia Redondo, Laura Mendoza-García, Carles Foguet, Juozas Grigas, Alba Calvet, José Manuel Villalba, María Josefa Rodríguez Gómez, Diego Megías, Biagio Mandracchia, Daniel Luque, Juan José Lozano, Cristina Calvo, Unai Merino Herrán, Timothy M. Thomson, Juan J. Garrido, Marta Cascante and María Montoya (2024) Journal of Medical Virology, 96:e29752. DOI:10.1002/jmv.29752

