A new work published in Blood journal by the group of Prof. Santiago Rodríguez de Córdoba at Centro de Investigaciones Biológicas Margarita Salas (CIB-CSIC) provides the functional characterization of Factor H variants associated with atypical hemolytic uremic syndrome (aHUS). This study provides a rationale for the classification of the FH variants found in the genetic screenings of patients with a clinical diagnosis of aHUS and, therefore, it aims to improve the medical use of the genetic information.
aHUS is a life-threatening thrombotic microangiopathy that can progress, when untreated, to end-stage renal disease. Most frequently, aHUS is caused by complement dysregulation due to pathogenic variants in genes that encode complement components and regulators. Amongst these genes, the Factor H (FH) gene presents with the highest frequency of variants (15-20%) and is associated with the poorest prognosis. Complement genetic testing is important to confirm aHUS diagnosis and to guide short- and long-term patient management with anti-complement therapeutics, but its clinical impact is limited by the large number of variants reported as 'variants of uncertain significance'. Classifying a FH variant as pathogenic or benign will not change the clinical diagnosis of aHUS. Instead, it will support (or exclude, if that is the only genetic finding in the patient) that constitutive complement dysregulation is the etiological factor underlying the development of aHUS. This information has crucial consequences for the clinical management of the patients.
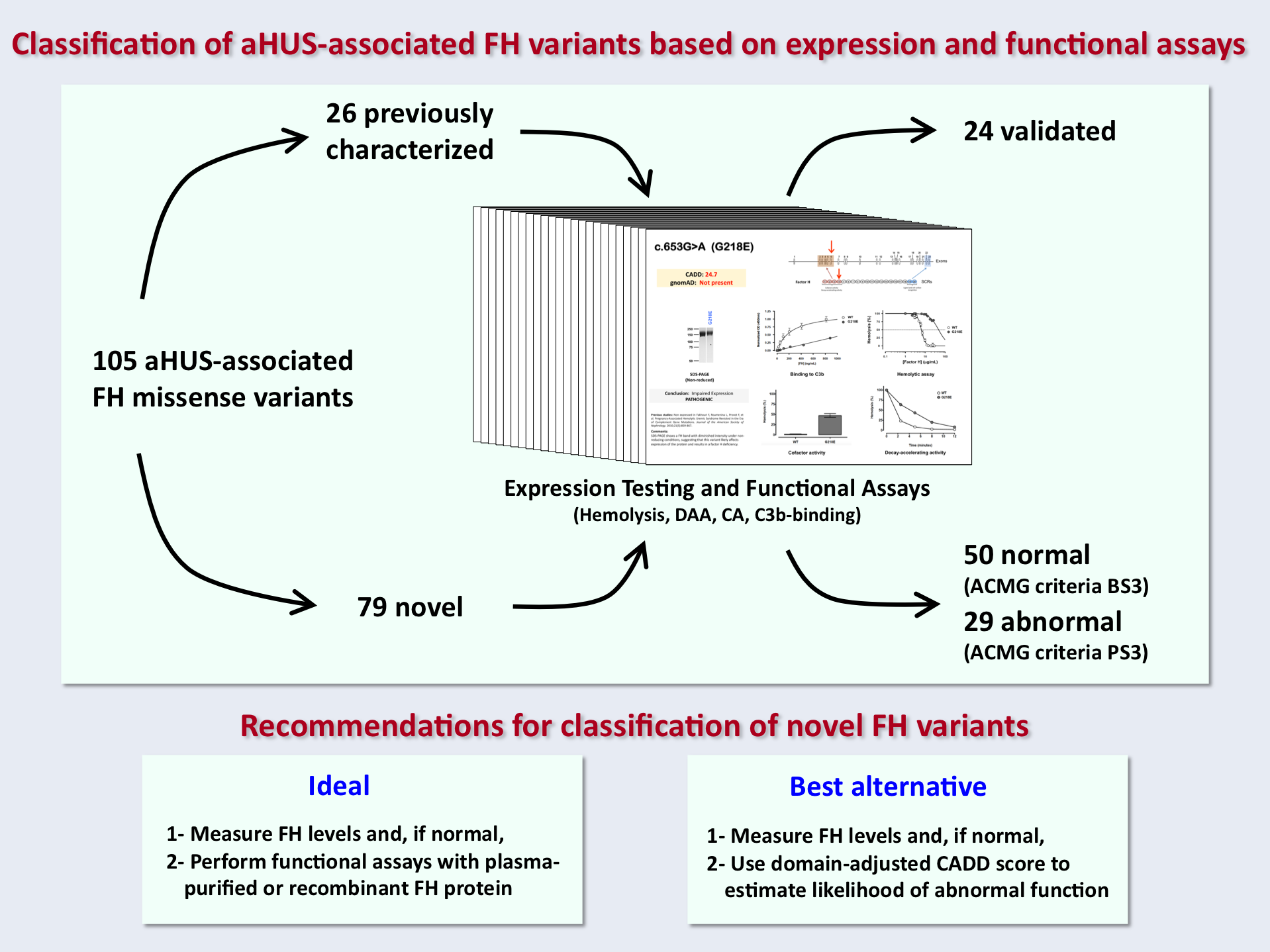
Martín-Merinero et al. expressed and functionally characterized 105 aHUS-associated FH variants. All FH variants were categorized as pathogenic or benign, and for each, the authors fully documented the nature of the pathogenicity. Twenty-six previously characterized FH variants were used as controls to validate and confirm the robustness of the functional assays used. Of the remaining 79 uncharacterized variants, only 29 (36.7%) alter FH in vitro expression or function and are therefore proposed to be pathogenic. The study highlights that rarity in control databases is not informative for variant classification, and identifies important limitations in applying prediction algorithms to FH variants.
The work remarks the need for functional assays to interpret FH variants accurately and offers ways to improve the prediction of variant classification when these technologies are not available.
Reference: Functional characterization of 105 Factor H variants associated with atypical HUS: lessons for variant classification. Hector Martin Merinero, Yuzhou Zhang, Emilia Arjona, Guillermo del Angel, Renee Goodfellow, Elena Gomez-Rubio, Rui-Ru Ji, Malkoa Michelena, Richard J. H. Smith, and Santiago Rodríguez de Córdoba (2021) Blood. DOI: 10.1182/blood.2021012037

