A recent investigation published in the journal Antioxidants and led by the researchers from the Margarita Salas Center for Biological Research (CSIC) Francisco Javier Ruiz-Dueñas and Ángel T. Martínez, identifies and characterizes for the first time a lignin peroxidase, responsible for the degradation of lignin, in a fungus of the order Agaricales, also showing that it is one of the most efficient ligninolytic enzymes described to date.
Lignin degradation is an essential process for carbon recycling in terrestrial ecosystems and often represents a key step for the use of plant biomass in industry. This process has traditionally been studied in the order Polyporales, which includes most of the fungi that degrade wood. The genomic analysis of these organisms and the characterization of their enzymes have highlighted the key role of fungal peroxidases with high redox potential (lignin peroxidases and others) in attacking lignin. However, the degradation of this aromatic polymer has hardly been studied in Agaricales, despite being the largest basidiomycete order, with around 13,000 described species, and representing a potential source of new lignocellulolytic enzymes given their ability to grow on a wide variety of lignocellulosic materials.
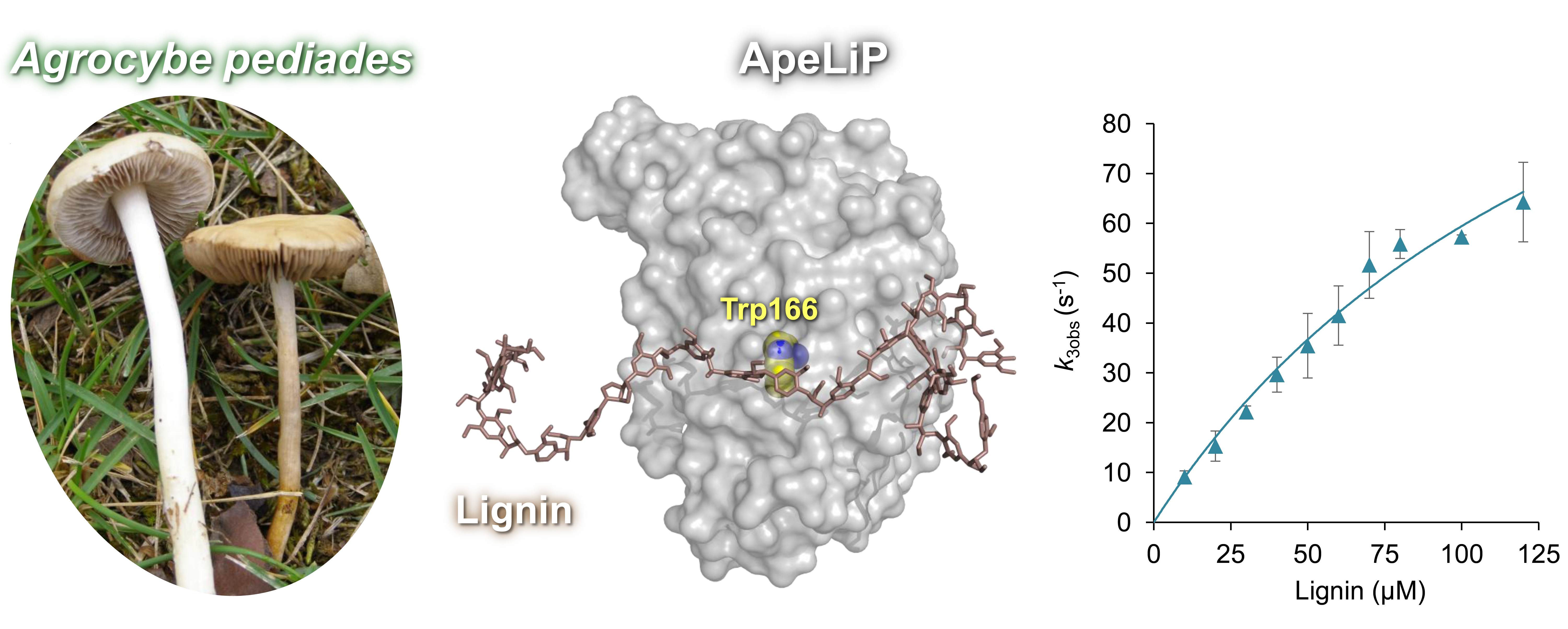
The investigation of dozens of genomes of Agaricales species carried out at the CIB Margarita Salas in collaboration with the Joint Genome Institute of the US Department of Energy (DOE-JGI) has revealed that lignin peroxidase genes appeared in this fungal order through an evolutionary pathway different from the one that gave rise to the Polyporales enzymes. Sánchez-Ruiz et al. have approached the study of the first of these peroxidases described in Agaricales, the lignin peroxidase from the fungus Agrocybe pediades (ApeLiP), whose genome has been recently sequenced within the framework of this collaborative project with DOE-JGI.
The study starts from the gene sequence of this enzyme and, using classical and cutting-edge techniques, including X-ray crystallography, site-directed mutagenesis, stopped-flow spectrophotometry, and two-dimensional nuclear magnetic resonance, delves into the structural and functional properties of the new peroxidase.
Its crystallographic structure, kinetic characterization, and the evaluation of its ligninolytic capacity reveal the catalytic mechanism of this enzyme, and show that it is not only capable of oxidizing model lignin compounds but also different types of real lignin, both from angiosperms and gymnosperms, in a degree similar or even higher than that of other known ligninolytic peroxidases.
This work shows that not only the wood-degrading Polyporales, but also the mushroom-forming Agaricales fungi, have enzymes of enormous relevance both for the recycling of carbon in nature and for the biotechnological modification of lignin.
The work is the result of collaboration between the groups of "Biotechnology for lignocellulosic biomass" and "Structural biology of proteins" of the CIB Margarita Salas, and the group "Vegetable biomass - Use and valorization" of the Institute of Natural Resources and Agrobiology of Seville (IRNAS).
Reference: Agaricales mushroom lignin peroxidase: from structure–function to degradative capabilities. Sánchez-Ruiz MI, Ayuso-Fernández I, Rencoret J, González-Ramírez AM, Linde D, Davó-Siguero I, Romero A, Gutiérrez A, Martínez AT, Ruiz-Dueñas FJ. Antioxidants (2021) 10:1446. https://doi.org/10.3390/antiox10091446

