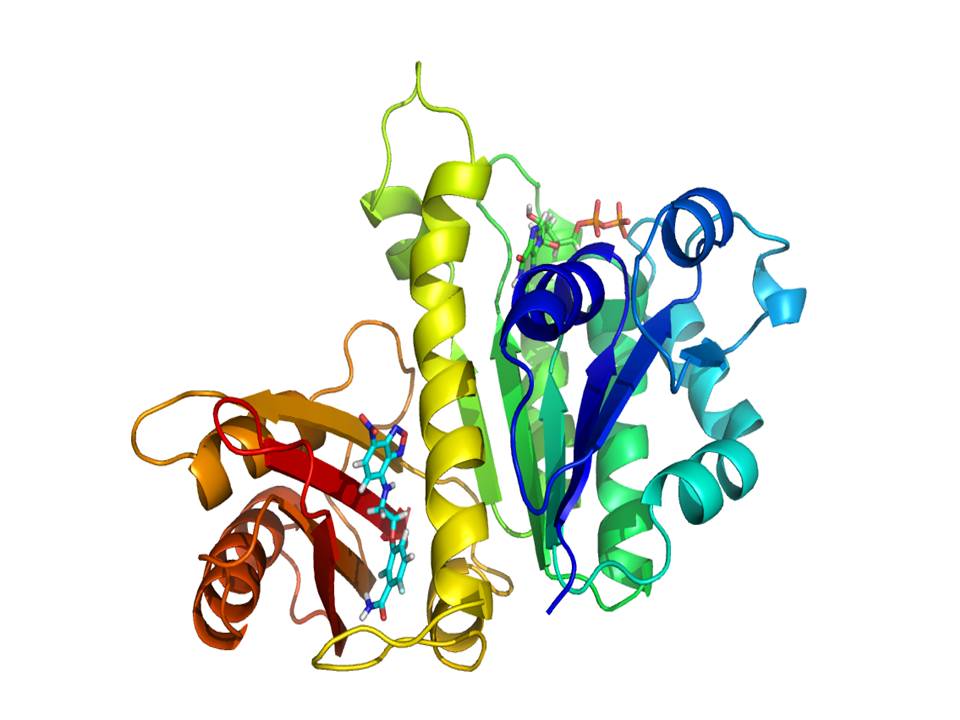
FtsZ is a widely conserved tubulin-like GTPase that directs bacterial cell division and a target for discovering new antibiotics needed to counter the spread of antimicrobial resistant pathogens. This fascinating protein assembly machine works cooperatively polymerizing into single-stranded filaments, by means of self-switching between inactive and actively associating monomer conformations. Structural comparisons had suggested a switch mechanism consisting of a cleft-opening movement of the C-terminal and N-terminal FtsZ domains, allosterically coupled to the formation of a tight association interface between consecutive subunits along the filament. A recent work published by the RSC journal Chemical Science (DOI: 10.1039/C6SC03792E) and carried out in collaboration between the CIB tubulins & FtsZ laboratory headed by José Manuel Andreu, the Medicinal Chemistry Lab from Universidad Complutense de Madrid and the Structural Bioinformatics Group from the CSIC Institute of Physical Chemistry Rocasolano has shown the FtsZ switch in action, as the cleft between both domains is open for assembled subunits and closed for unassembled monomers in the solution. To this purpose we synthesized fluorescent analogs of the antibacterial FtsZ inhibitor PC190723, whose fluorescence anisotropy increases upon specific binding to the open cleft in polymers of FtsZ from Staphylococcus aureus. We have also found that these probes directly label FtsZ division rings in live bacterial cells. We envisage that competitive fluorescent assays combined with cytological profiling methods may be employed to screen for allosteric inhibitors of FtsZ assembly in search of antibiotics with new mechanism of action.

